ABSTRACT: A new commercial capillary column containing both strong cation exchange (SCX) and reversed phase (RP) supports is now available. The column can be used to automate MudPIT-style separations of peptides from complex samples on a single-dimensional HPLC. A novel method employs pH steps with MS-compatible buffers to elute peptides from the SCX region of the column. Eluted peptides flow to the adjacent C18 and are separated by capillary reversed phase HPLC. This method has been used to identify thousands of proteins in tryptic digests of mouse liver lysates.
INTRODUCTION: To analyze a complex protein sample, the method of "shotgun sequencing" is often employed. A sample is digested with a suitable protease, commonly trypsin, and the resulting peptides are separated by two-dimensional (2D) HPLC and characterized by tandem mass spectrometry (MS). Proteins are identified by matching the observed MS/MS fragmentation pattern of their peptides with patterns predicted by information from genomic or proteomic databases.
Today, the most prevalent 2D HPLC technique employs a combination of SCX and C18 HPLC. By creating on-line SCX "fractions" this method greatly reduces the complexity of the peptide mixture being analyzed by the mass spectrometer at any given time point in time, improving the odds that a particular peptide will be detected. This is especially true for less abundant or poorly ionized peptides.
The major limitations of this technique have been: 1) complications with MS created by salt elution of peptides from the SCX support and 2) a lack of robust commercial columns for this application, requiring that researchers pack their own bi-phasic capillary columns and 3) a requirement for expensive, complicated multi-dimensional LC systems to properly execute these techniques.
In this paper we describe a novel, simplified, integrated bi-phasic column method for multi-dimensional LC-MS/MS protein analysis. A single capillary column, packed with both SCX and C18 supports is used. To avoid the use of salts that will suppress peptides during LC/MS analysis, peptides are eluted from the SCX support with pH steps or gradients formed from MS-compatible buffers. Peptides are retained on the SCX support until exposed to a pH at or above their pI, at which point they move to the C18 resin. Using this method, there is no need to use any kind of salt and no need for extensive column washing. The pH buffers can be injected by an autosampler which allows use of a single-pump HPLC system to achieve multi-dimensional chromatographic separations.
EXPERIMENTAL: The sample was a highly complex peptide mixture created by tryptic digestion of a mouse liver proteome lysates.
The sample was analyzed by bi-phasic LC-MS/MS, using a Surveyor™ high-performance liquid chromatography system with autosampler and one high pressure HPLC pump (Thermo Electron, San Jose, CA) coupled with a Finnigan™ LTQ™ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). A PepSil™ Bi-Phasic Column, with both SCX and C18 supports, (5 cm x 320µm + 10 cm x 150µm, Column technology Inc., Fremont, CA Part#: CTIBiphase) was used. This column is shown in Figure 1. The RP HPLC solvents used were 0.1% formic acid (v/v) aqueous (A) and 0.1% formic acid (v/v) acetonitrile (B).
600µg of sample was loaded and washed with 5% solvent B. For each pH step, 100 µl of pH buffer was injected with the autosampler and the column was washed with 5% solvent B. The total injection and wash time for each pH step was about 80 minutes at a flow rate of 300 µl/min. With each pH step, a fraction of peptides were eluted from the SCX section at the head of the bi-phasic column to the reversed phase body of the column. A reversed phase gradient was run, from 5 to 65% solvent B in 115 minutes at 200 µl/min. This procedure was followed for each of nine pH steps: 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, and 8.0. ABi-Phasic Buffer Kit (Column technology Inc., Fremont, CA Part#: CTIBiphaseBuffers) was used to create the pH steps.
The Finnigan LTQ was equipped with an electrospray interface (Thermo). The temperature of ion transfer capillary was set at 160ºC. A voltage of 3.0 kV was applied to the ESI needle and the normalized collision energy was 35.0%. All MS and MS/MS spectra were acquired in the data-dependent mode. The mass spectrometer acquired one full MS scan, followed by ten MS/MS scans on the ten most intense ions from the MS spectrum. System control and data collection were done by Xcalibur software version 1.4 (Thermo). Proteins were identified with SEQUEST using the following criteria for acceptable protein ID: Xcorr > 1.9, 2.2, 3.75 for +1, +2, +3 charged peptides; Cn < 0.1.
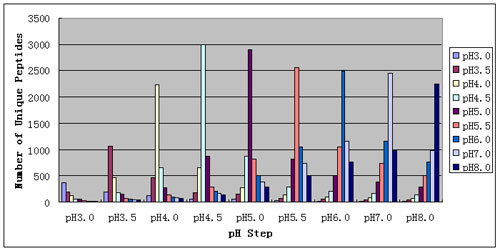
RESULTS AND DISCUSSION: The system described was able to identify 5779 unique peptides and 2108 proteins in the mouse liver proteome. This represents a significant increase over previous 1D RP-HPLC methods (data not shown) and multi-column 2D HPLC (data not shown) analyses of this sample. As seen in Figure 2, pH effectively fractionated the sample into groups of peptides that were efficiently resolved by RP HPLC. As seen in Figure 3, each pH step eluted many unique peptides and most peptides eluted in a single pH step. Most peptides eluted from pH 3.5 to 5.5, in good agreement with theoretical tryptic peptide pI calculations.
Analysis of complex proteomic samples by data dependent MS/MS is practically limited by the resolving power of the separation system and the sensitivity and cycle time of the mass spectrometer. The Finnigan LTQ is extremely sensitive and has a very rapid cycle time, typically 0.2s/full scan MS/MS spectrum. However, even this extremely capable system is quickly overwhelmed when presented by a large number of co-eluting peptides, as will be the case in complex proteomic analyses. To achieve the highest possible number of peptide and protein identifications, peptides must be presented to the mass spectrometer in manageable numbers – the sample must be separated into the largest number of peaks possible. In other words, the peak capacity of the system must be maximized. This can only be achieved by a combination of fractionation and high resolution separation. These methods can be complex and time consuming.
Figure 1: PepSil Bi-Phasic Column. The first 5cm of the column is packed with PepSil SCX resin. The remaining 15cm of the column is packed with PepSil C18.

Figure 2: Peptide fractions are created by pH elution of the SCX packing material at the head of the PepSil Bi-Phasic Column. Each of these fractions are then separated by the reversed phase C18 material in the body of the column. Total Ion Chromatograms (TIC) are shown for each of these steps. The whole procedure can be performed with a single high pressure HPLC pump and is completely automated.
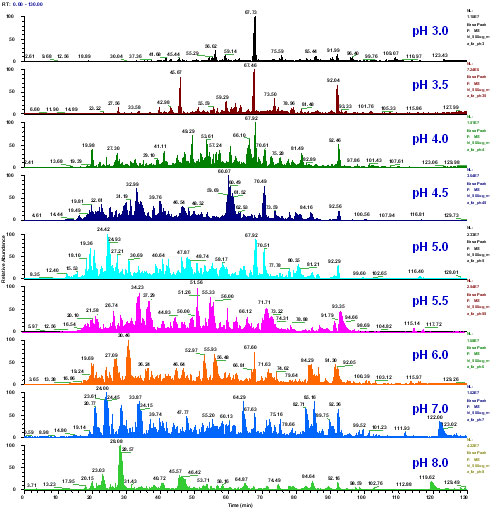
Figure 3: shows the overlap of peptides across pH steps. Such overlap is often caused by mixed-mode interaction of peptides with an SCX column. The PepSil Bi-Phasic Column displays very low overlap with almost all peptides specifically eluting in a single pH step. In comparison, with methods that use salt elution, the same peptide will be seen in many salt steps, reducing the overall peak capacity of the system (data not shown). Most peptides eluted at pH 4-5 which agrees with theoretical tryptic peptide pI calculations.